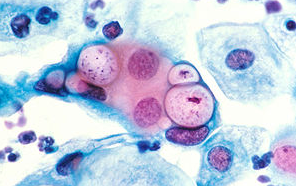
클라미디아 감염의 원인과 예방 방법
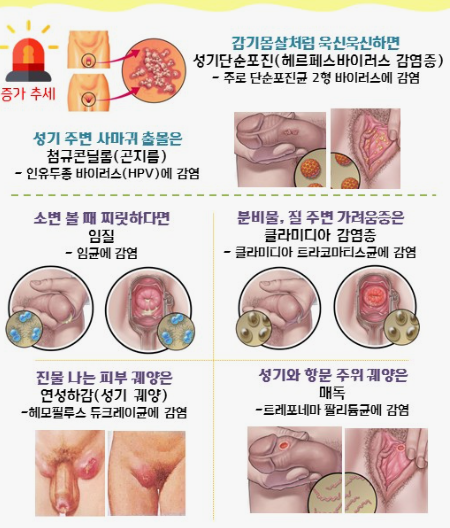
클라미디아는 성병으로서, 세계적으로 많은 사람들에게 영향을 미치고 있는 질병 중 하나입니다. 이러한 감염이 발생한 이유에는 여러 가지 요인들이 작용하고 있습니다. 오늘은 클라미디아 감염이 생겨난 이유와 예방 방법에 대해 알아보겠습니다. 1. 성접촉을 통한 전파 클라미디아는 성적 접촉을 통해 전파되는 성병 중 하나입니다. 감염된 사람과의 성관계를 통해 성기간에 감염될 수 있습니다. 특히, 무보호 성관계를 하거나 여러 파트너와 […]
계속 읽기